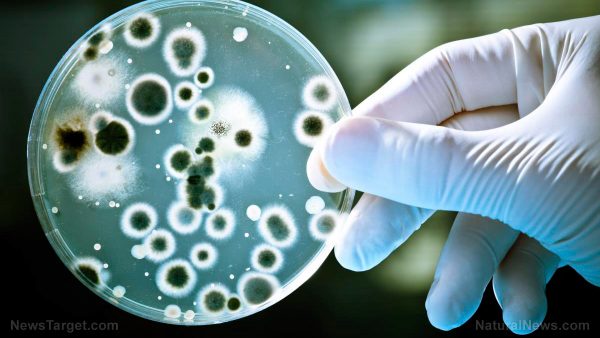
Hospital pipes, full of DNA fragments, found to be a breeding ground of antibiotic resistance
08/27/2018 / By Ralph Flores

Antibiotic resistance is a growing cause for concern, especially in cases where it happens in hospitals or other healthcare facilities. To better understand how these organisms spread in this kind of environment, researchers from the National Institutes of Health (NIH) collected samples from various areas of the hospital to investigate, allowing them to identify where these organisms are found and how they spread in a hospital setting. The results, which were published in the American Society for Microbiology’s open access journal mBio, indicated that plasmids that provide resistance to carbapenem – a last-line-of-defense antibiotic used against multi-drug-resistant organisms – were found in the hospital’s plumbing.
Carbapenems are part of the beta-lactam types of antibiotics. Together with penicillin, cephalosporin, and monobactam, they bind and inactivate penicillin-binding proteins, which are found in the cell wall. Of the beta-lactams, carbapenems present one of the broadest range of antibacterial activity. This makes them a reliable last-resort treatment for antibiotic-resistant strains.
The amount of antibiotic-resistant organisms found in the wastewater can be attributed to the heavy use of antibiotics in hospitals, researchers posited in the study. This results in an environment where microorganisms fight to survive, according to NIH microbiologist Karen Frank. “The bacteria fight with each other and plasmids can carry genes that help them survive,” she says.
The NIH’s Hospital Epidemiology Service regularly conducts an extensive sampling of high-touch surfaces, sinks, and other areas in the hospital; however, for this study, the range was expanded to cover housekeeping closets, wastewater from hospital internal pipes, and external manholes over a two-year period. Moreover, the team collected five years’ worth of patient clinical and surveillance isolates, and this was compared with the samples collected. A total of 108 isolates underwent whole-genome sequencing and analysis to enable an in-depth genetic comparison.
Tests revealed that wastewater from pipes that drain out of the intensive care unit and external manholes had carbapenemase-producing organisms (CPOs). These organisms were made up of different species and plasmids, which could have differing effects on the environment and people. Also, plasmid backbones common to both populations were also detected. This, according to the investigators, indicates a potentially large supply of factors that are conducive to the spread of antibiotic-resistant genes. (Related: Researchers identify natural weapons in the fight against antibiotic resistance.)
“The finding leads one to consider what might be found if more hospitals were investigated to this extent,” the researchers wrote. “It is likely that most hospitals have some carbapenemase-producing organism colonization in wastewater and drains that remain undetected.”
The results from the sequence analysis and epidemiology indicate that infection control and compliance measures were successful. However, researchers found a case of Leclercia sp. that was transmitted from the hospital in the surveillance.
Still, Frank thinks it should not alarm the general public. “I would say it is not a huge concern in the United States,” she said. “Healthy people tend to be resistant against this.” She adds, however, that high-risk patients, such as those in intensive care or those with compromised immune systems, may still be prone to infections.
The results of the investigation, according to the researchers, improve the understanding of how CPOs are transmitted in a hospital environment and could be useful in future infection control strategies. As for hospitals, the NIH recommends changing their cleaning practices.
“They can pay attention to the particular cleaning agents that are used,” Frank added. “We upgraded our cleaning agents,” she said.
Learn more about antibiotic-resistance and how to avoid it by heading to Superbugs.news today.
Sources include:
Tagged Under: antibiotic resistance, Antibiotics, carbapenem, carbapenemase-producing organisms, hospital germs, Hospitals, infection control, infection control strategies, infectious disease, intensive care unit, Leclercia sp., manholes, multidrug-resistant organisms, plasmids, research, superbugs, wastewater, whole-genome sequencing